On
my GitHub page, you can find
repository with Python codes related to my computational research on intrinsically disordered protein (IDP) phase separation. These codes implement a field-theoretic model of polyampholytes interacting through long-range electrostatic forces and short-range excluded volume. This is a simple but widely studied model that captures many features of electrostatically driven IDP phase separation.
Key scripts include:
-
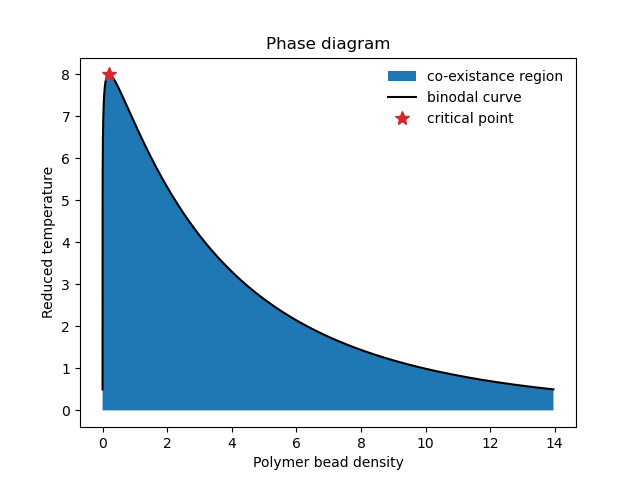
RPA_polyampholytes.py: This code can be used for computing the coexistence curve (binodal) in the random phase approximation (RPA).
-
FTS_polyampholytes.py: This code defines the basic object and functions for fully fluctuation field-theoretic simulations (FTS).
-
FTS_trajectories_MPI.py: This code can be used for running parallelized FTS to compute the chemical potential and osmotic pressure at different bulk polymer densities.
-
FTS_analyze_trajectories.py: This script analyzes the output files from FTS_trajectories_MPI.py to compute the coexistence curve.
A detailed user guide is provided in this
PDF. The codes in this repository are published as a part of the following book chapter, wherein the theory, methods and key references are provided:
-
Numerical Techniques for Applications of Analytical Theories to Sequence-Dependent Phase Separations of Intrinsically Disordered Proteins
Y.-H. Lin*, J. Wessén*, T. Pal*, S. Das and H. S. Chan (*equal contribution)
In: Zhou, HX., Spille, JH., Banerjee, P.R. (eds) Phase-Separated Biomolecular Condensates (2023). Methods in Molecular Biology, vol 2563. Humana, New York, NY.
DOI: doi.org/10.1007/978-1-0716-2663-4_3
Preprint: arXiv:2201.01920